Introduction
descs-standalone is a tool allowing a user to identify and structurally compare local, contact-based structural motifs, called descriptors. A comprehensive description of the proposed tool and provided algorithms is given in the article published in BMC Bioinformatics. The descriptors can be built on unmodified residues of biological molecules such as proteins and RNAs. Both PDB and CIF formats are supported. At the beginning of the processing, a comprehensive validation of the input tertiary structures is performed. As a result, all identified inconsistencies are filtered out and stored in a log file.
Features
Features of the tool include:
-
Identification of descriptors observed in the structural neighborhood of every residue of the input 3D structure of a molecule.
-
» A flexible representation of an expression used for an identification of in-contact residues located in the proximity of the descriptor's center, that can be simply introduced by the user.
The tool supports basic operators: logical (i.e., OR, AND, NOT), relational (i.e., <, <=, =, >=, >) and arithmetic ones. A user can introduce the DISTANCE operator between any atoms, except hydrogens, that are found in the 3D structure of the input molecule (e.g., DISTANCE:C1';O5', DISTANCE:CA). Moreover, several virtual atoms can be also applied, i.e., in proteins: geometric centers of a backbone [BBGC] and a side chain [SCGC], CB extended point [CBX], and virtual CB atom provided by biojava [VCB], while in RNAs: geometric centers of a backbone [BBGC], a ribose [RBGC] and a base [BSGC].
An example expression is presented below:
OR(DISTANCE:SCGC < = 6.5, AND(DISTANCE:SCGC < = DISTANCE:CA - 0.75, DISTANCE:SCGC <= 8.0))
- » The size of the descriptor element can be configured by the user.
- » The output descriptor set can be constrained by the user through thresholds associated with the number of segments, elements and residues.
- » A concurrent processing is supported to increase processing efficiency, the number of threads can be configured by the user.
Information about either the central residue or other residues being in-contact with the descriptor center, and the number of adjacent residues on each side of the element center are stored in the following fields of the coordinate section, namely temperature factor and occupancy, respectively.
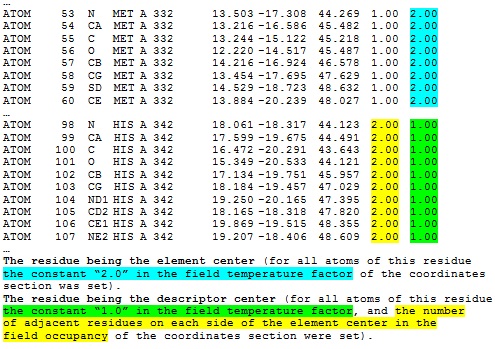
Figure 1. 3D structure template of a descriptor (assumed length of an element is five residues).
-
-
Structural comparison of a descriptor pair performed with the use of several computationally efficient algorithms.
- » Backtracking-driven exact algorithms.
- » Hungarian method-driven heuristic algorithms.
- » Thresholds (i.e., a maximal RMSD of the pair of aligned central elements, a maximal RMSD of a pair of aligned duplexes, a minimal fraction of aligned elements, a minimal fraction of aligned residues, a maximal RMSD of the total alignment) driving a multi-criteria function of the structural similarity of descriptors can be flexibly configured by the user.
- » Acceptance criteria, used for identification of a potentially better alignment, can be chosen by the user (i.e., ALIGNED_RESIDUES_ONLY, ALIGNED_RESIDUES_AND_AVERAGE_RMSD_OF_ALIGNED_DUPLEXES).
- » A result of the comparison can be complemented with 3D structures of the aligned descriptors.
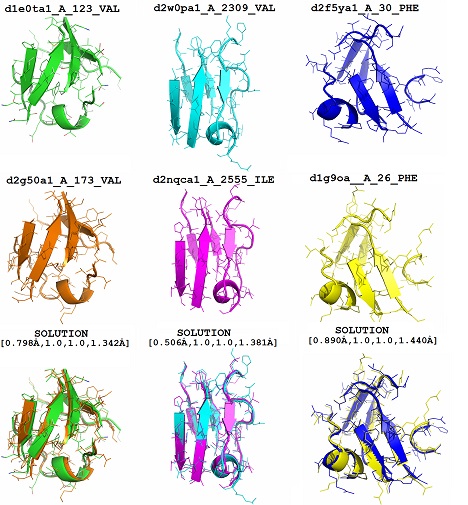
Figure 2. Visualization of example instances of structural comparison of protein descriptors and the corresponding optimal solutions. First two rows present descriptors that are compared and the third one is for their optimal structural alignment.
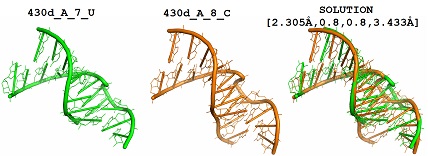
Figure 3. Visualization of an example instance of structural comparison of RNA descriptors and the corresponding optimal solution. First two columns present descriptors that are compared and the third one is for their optimal structural alignment.
-
Format conversion of tertiary structures of considered biological molecules from PDB to CIF and vice versa.
- » The support for generation of EBI-inspired, compatible PDB file bundles (tar.gz) in the case of conversion of 3D structures of large biomolecules that are stored in format CIF only.
Execution modes
The tool provides the following execution modes: FORMAT_CONVERSION, DESCRIPTORS_BUILDING, DESCRIPTORS_COMPARISON that can be set with one of the options -em,–execution-mode (default=FORMAT_CONVERSION).
- » execution-mode = FORMAT_CONVERSION
-i,--input-file <arg> input file path
-if,--input-format <arg> supported file formats: PDB, CIF
-o,--output-file <arg> (optional) output file path
-of,--output-format <arg> supported file formats: PDB, CIF
- » execution-mode = DESCRIPTORS_BUILDING
-es,--element-size <arg> (optional) number of residues in a single element
[default=5]
-fecge,--filter-of-descriptors-that-characterized-with-lower-value-of-elements-count
<arg> (optional) a filter on descriptors that are characterized by lower
value of elements count than the given bound [default=1]
-fecle,--filter-of-descriptors-that-characterized-with-higher-value-of-elements-count
<arg> (optional) a filter on descriptors that are characterized by higher
value of elements count than the given bound [default=200]
-frcge,--filter-of-descriptors-that-characterized-with-lower-value-of-residues-count
<arg> (optional) a filter on descriptors that are characterized by lower
value of residues count than the given bound [default=1]
-frcle,--filter-of-descriptors-that-characterized-with-higher-value-of-residues-count
<arg> (optional) a filter on descriptors that are characterized by higher
value of residues count than the given bound [default=1000]
-fscge,--filter-of-descriptors-that-characterized-with-lower-value-of-segments-count
<arg> (optional) a filter on descriptors that are characterized by lower
value of segments count than the given bound [default=1]
-fscle,--filter-of-descriptors-that-characterized-with-higher-value-of-segments-count
<arg> (optional) a filter on descriptors that are characterized by higher
value of segments count than the given bound [default=50]
-i,--input-file <arg> input file path
-ice,--in-contact-residues-expression-file <arg> file path of the expression that
should be fulfilled by each in-contact residues pair
-if,--input-format <arg> (optional) supported file formats: PDB, CIF [default=PDB]
-mt,--molecule-type <arg> supported molecule types: PROTEIN, RNA
-od,--output-directory <arg> (optional) output directory path
-of,--output-format <arg> (optional) supported file formats: PDB, CIF [default=PDB]
-tc,--threads-count <arg> (optional) number of threads used during processing
[default=AVAILABLE_PROCESSING_UNITS_COUNT]
- » execution-mode = DESCRIPTORS_COMPARISON
-aam,--alignment-acceptance-mode <arg>
(optional) alignment acceptance mode, supported modes: ALIGNED_RESIDUES_ONLY,
ALIGNED_RESIDUES_AND_AVERAGE_RMSD_OF_ALIGNED_DUPLEXES
[default=ALIGNED_RESIDUES_AND_AVERAGE_RMSD_OF_ALIGNED_DUPLEXES]
-aan,--file-path-of-atom-names-used-during-alignment-building <arg>
file path of atom names considered during building the alignment
-cat,--comparison-algorithm-type <arg> type of the comparison algorithm:
BACKTRACKING_DRIVEN_LONGEST_ALIGNMENT,
BACKTRACKING_DRIVEN_FIRST_ALIGNMENT_ONLY,
HUNGARIAN_METHOD_DRIVEN_FIRST_ALIGNMENT_ONLY_PARTIAL_SOLUTIONS_NOT_CONSIDERED,
HUNGARIAN_METHOD_DRIVEN_LONGEST_ALIGNMENT_PARTIAL_SOLUTIONS_NOT_CONSIDERED,
HUNGARIAN_METHOD_DRIVEN_LONGEST_ALIGNMENT_PARTIAL_SOLUTIONS_CONSIDERED <arg>
-fd,--file-path-of-first-descriptor <arg> file path of the first descriptor
-if,--input-format <arg> (optional) supported file formats: PDB, CIF [default=PDB]
-maep,--minimal-fraction-of-aligned-elements <arg>
(optional) minimal fraction of aligned elements [default=4/5]
-magrmsd,--maximal-rmsd-of-total-alignment <arg>
(optional) maximal RMSD of the total alignment [default=3.5A]
-marp,--minimal-fraction-of-aligned-residues <arg>
(optional) minimal fraction of aligned residues [default=2/3]
-mdparmsd,--maximal-rmsd-of-pair-of-aligned-duplexes <arg>
(optional) maximal RMSD of a pair of aligned duplexes [default=3.5A]
-moeparmsd,--maximal-rmsd-of-central-elements-alignment <arg>
(optional) maximal RMSD of the central elements alignment [default=1.2A]
-mrmsdtpdp,--maximal-cost-of-pair-of-aligned-duplexes <arg>
(optional) threshold f used by the Hungarian method-driven heuristics
as a maximal average cost per a pair of aligned duplexes [default=2.33]
-mt,--molecule-type <arg> supported molecule types: PROTEIN, RNA
-od,--output-directory <arg> output directory path
-of,--output-format <arg> (optional) supported file formats: PDB, CIF [default=PDB]
-sd,--file-path-of-second-descriptor <arg> file path of the second descriptor
-wa,--with-alignment <arg> (optional) a result of the comparison can be complemented
with 3D structures of aligned descriptors, supported modes: CONSIDER, IGNORE
[default=IGNORE]
How to run descs-standalone
-
descs-standalone binaries and usage scenario examples can be downloaded from here (39.8 MB).
-
To run descs-standalone one must have installed:
- » stable release of Oracle JDK 6 or above (however, Oracle JDK 7 is recommended).
A used version of Java can be configured by setting the JAVA_HOME environment variable.
How to build descs-standalone
-
descs-standalone is available as an open source project stored on

-
To build descs-standalone one must have installed:
- » stable release of Oracle JDK 6 or above (however, Oracle JDK 7 is recommended),
- » stable release of Apache Maven 3.0.3 or above,
- » stable release of Git.
A used version of Java can be configured by setting the JAVA_HOME environment variable.
Data sets
In computational experiments the following data sets were used:
-
The set of tertiary structures of selected protein domains, provided by the ASTRAL compendium for protein structure, that was used in the generation of descriptors experiment can be downloaded from here (66.8 MB).
-
The set of tertiary structures of selected protein descriptors consisting of at least three segments, that was used in the structural comparison of descriptors experiment can be downloaded from here (52.1 MB).
Acknowledgements
We thank Prof. Krzysztof Fidelis and Andriy Kryshtafovych from the Protein Structure Prediction Center, UC Davis Genome Center, for valuable cooperation, sharing of ideas and discussions.
Funding
The research was supported by the National Science Centre, Poland [grant No. 2012/05/B/ST6/03026].
How to cite descs-standalone
Antczak, M., Kasprzak, M., Lukasiak, P., Blazewicz, J., Structural alignment of protein descriptors - a combinatorial model, BMC Bioinformatics, 2016, 17:383, (doi:10.1186/s12859-016-1237-9) [PDF].
Contact us
If you have any questions, comments or suggestions you can contact us by sending electronic mail to Maciej Antczak.
